Overview
The field of Life Sciences is grappling with an explosion of data. This crucial information, such as spanning research papers, clinical trial reports, patient records, and even genomic sequences, exists as unstructured text. Transforming this vast textual landscape into actionable insights is a significant challenge. This is where the power of Natural Language Processing (NLP) and especially Named Entity Recognition (NER) comes into play.
Natural Language Processing is a discipline within Artificial Intelligence (AI) that focuses on building machines capable of manipulating human language. In recent years, NLP has greatly improved – not only in understanding human language, but also in reading patterns in things like DNA and proteins, which are structured like language.
Named Entity Recognition (NER)
The following diagram illustrates the NER process in detail.
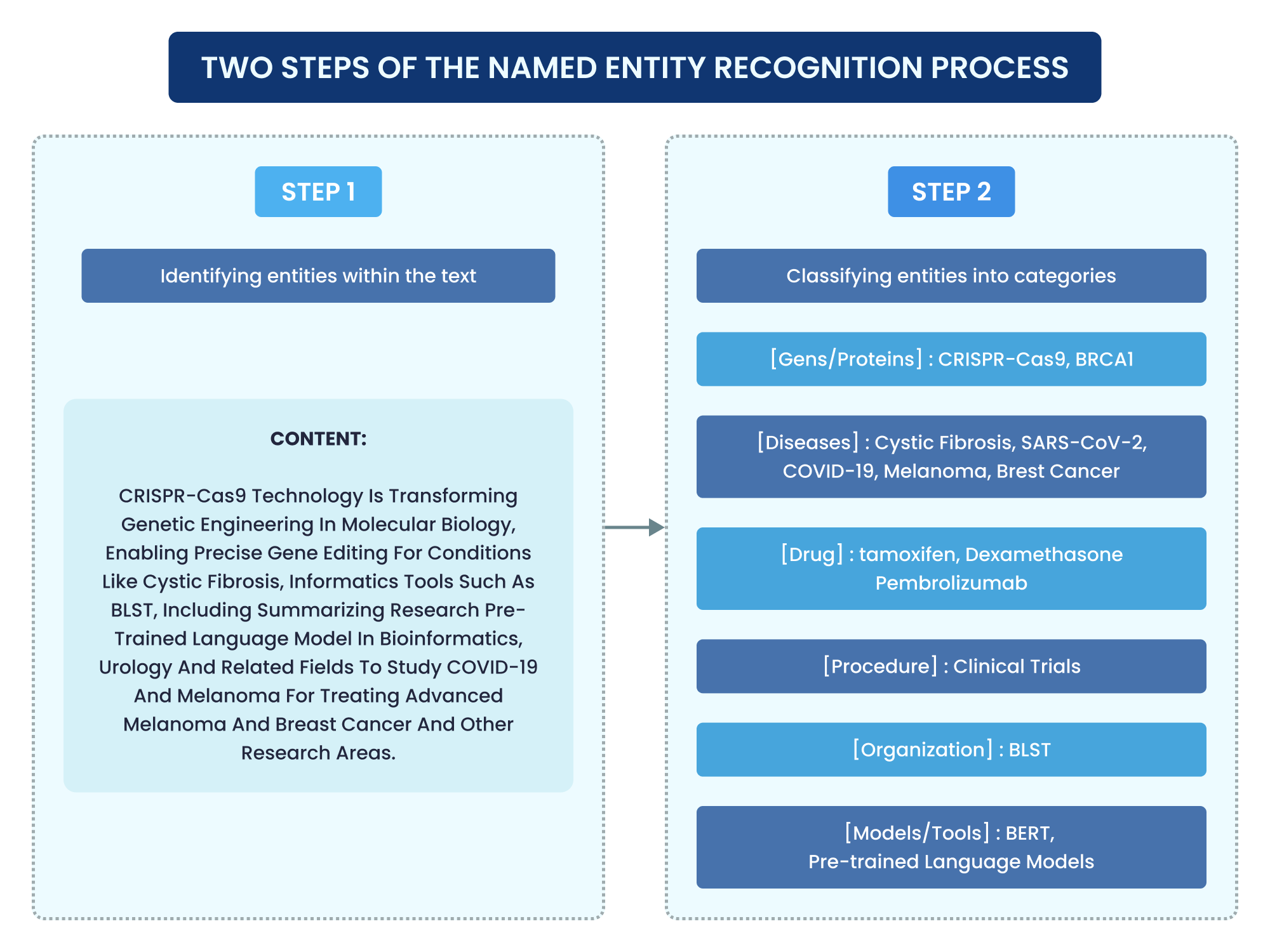 Named Entity Recognition is an indispensable technique in NLP. Think of NER as a wizard that sifts through text to find and categorize specific “treasures” – named entities. It’s a sub-task of information extraction. NER goes beyond simple word labeling and assigns contextually relevant entity types to words or subwords.
Named Entity Recognition is an indispensable technique in NLP. Think of NER as a wizard that sifts through text to find and categorize specific “treasures” – named entities. It’s a sub-task of information extraction. NER goes beyond simple word labeling and assigns contextually relevant entity types to words or subwords.
Its primary purpose is to comb through unstructured text, identify specific chunks as named entities, and subsequently classify them into predefined categories. These categories commonly include person names, organizations, locations, dates, monetary values, quantities, and time expressions. Notably for Life Sciences, predefined categories can also include medical codes. By converting raw text into structured information, NER facilitates tasks like data analysis, information retrieval, and knowledge graph construction.
Consider the sentence: “J & J received FDA approval for its COVID-19 vaccine, Janssen, in the United States in 2021.” Using the NER principles described in the sources, an NER system would process this sentence.
How NER Works: A Step-by-Step Process
The process of NER, while complex, can be broken down into several key steps:
- Tokenization: The initial step involves dividing the text into smaller units called tokens, which can be words, phrases, or even sentences. For instance, “J & J”, “received”, “FDA”, “approval”, “for”, “its”, “COVID-19”, “vaccine”, “,”, “Janssen”, “,”, “in”, “the”, “United”, “States”, “in”, “2021”,
- Feature Extraction / Entity Identification: Linguistic features such as part-of-speech tags, word embeddings, and context are extracted for each token. Alternatively, potential named entities are detected using linguistic rules, regular expressions, dictionaries, or statistical methods. This involves recognizing patterns like capitalization (“Steve Jobs”) or specific formats.
- Entity Identification and Classification: The system identifies potential entities and classifies them into predefined categories. Drawing on the types of entities NER handles and extending them for a healthcare/pharma domain (which often involves specific products and medical conditions), NER would likely identify:
- “J & J” as an ORGANIZATION. This aligns directly with the “organizations” category mentioned in the sources.
- “FDA” (Food and Drug Administration) as another ORGANIZATION. This is also a type of organization that NER would classify.
- “COVID-19” as a DISEASE or MEDICAL CONDITION. While “medical codes” are mentioned, a system tuned for this domain would likely have a specific category for diseases, drawing on the concept of identifying “more” entity types beyond the standard list.
- “Janssen” as a PRODUCT or DRUG. This would also be a domain-specific category relevant to pharmaceuticals, extending the core entity types to capture specific items of interest in the industry, akin to identifying products in customer support analysis.
- “United States” as a LOCATION. This aligns directly with the “locations” category.
- “2021” as a DATE. This aligns directly with the “dates” category.
- Entity Span Identification: Beyond classification, NER also identifies the exact beginning and end of each entity mention within the text. This is crucial for precise data extraction.
- Contextual Understanding / Contextual Analysis: Modern NER models are sophisticated enough to consider the surrounding text to improve accuracy. For example, the context in “J & J released a new vaccine” helps the system recognize “J & J” as a company. Models like BERT and RoBERTa use contextual embeddings to capture word meaning based on context, helping handle ambiguity and complex structures.
- Post-processing: After the initial steps, post-processing is applied to refine the results. This can involve resolving ambiguities, merging multi-token entities (like “New York” being a single location entity), or using knowledge bases for richer entity data.
The power of NER lies in its capacity to understand and interpret unstructured text, adding structure and meaning to the vast amount of textual data we encounter.
Beyond NER: Advanced NLP Techniques
While NER is fundamental, Life Sciences often require a more sophisticated understanding of language. Advanced NLP techniques, many empowered by deep learning, enable complex tasks that complement NER.
Information Extraction: NER is a key component, but Information Extraction extends to extracting structured information (like relationships between entities) from unstructured text to populate databases or build knowledge graphs.
Question Answering (QA): Systems can identify entities in user queries (using NER) and find relevant answers in documents. QA systems can be multiple-choice or open-domain, providing answers in natural language.
Summarization: This task shortens text while retaining key information. Extractive summarization pulls key sentences, while Abstractive summarization paraphrases, potentially using words not in the original text. This is useful for condensing research papers or clinical notes.
Topic Modeling: An unsupervised technique that discovers abstract topics within a corpus of documents. It views documents as collections of topics and topics as collections of words (like Latent Dirichlet Allocation – LDA). This can identify prevalent research themes.
Sentiment Analysis: Classifies the emotional intent of text (positive, negative, neutral). Understanding sentiment associated with entities identified by NER can provide deeper insights. This could be applied to patient feedback or social media discussions about treatments.
Text Generation (NLG): Produces human-like text. While less directly tied to the analysis of existing Life Sciences text, advanced models can generate drafts of reports or summaries.
Information Retrieval: Finds documents most relevant to a query that are crucial for searching vast literature databases.
Why Life Sciences Needs NLP and NER
Life Sciences is drowning in data, most of which is locked within unstructured text documents. NLP and NER are crucial because they provide the means to:
Transform Unstructured Data: They serve as a bridge, converting vast amounts of raw textual information into structured, categorized forms that machines can easily process and analyze.
Accelerate Research & Discovery: Researchers can rapidly scan massive volumes of literature, identifying mentions of specific entities (genes, proteins, diseases) relevant to their studies, speeding up data analysis.
Improve Clinical Care: Interpreting or summarizing complex electronic health records (EHRs) becomes feasible. Extracting key information like patient history, symptoms, treatments, and outcomes can enhance decision-making. NER can potentially identify medical codes or other critical entities within these records.
Enhance Knowledge Management: Building knowledge graphs by identifying entities and their relationships from scientific literature or clinical data is facilitated by NER and information extraction.
Support Compliance and Analysis: Automating the tedious process of sifting through legal or regulatory documents to find relevant information becomes possible.
Analyze Biological/Chemical Sequences: Some NLP techniques, like those dealing with data resembling language, can potentially be applied to analyzing biological sequences.
Leveraging NER and Advanced NLP: Use Cases in Life Sciences
Based on the capabilities described in the sources, here are some potential applications within the Life Sciences domain:
Biomedical Entity Recognition: Identifying and classifying entities specific to Life Sciences, such as genes, proteins, diseases, drugs, chemical compounds, and procedures from research papers, patents, or clinical text. This leverages the core NER capability for domain-specific entities.
Relationship Extraction from Literature: Automatically identifying relationships between biomedical entities mentioned in research articles, e.g., drug-gene interactions, disease-symptom associations, protein-protein interactions. This builds upon Information Extraction techniques facilitated by NER.
Clinical Text Analysis: Extracting structured information from clinical notes, discharge summaries, and other EHR components, including patient demographics, symptoms, diagnoses, medications, lab results, and treatment plans. NER identifying medical codes could be a key part of this.
Summarizing Scientific Literature and Clinical Trials: Automatically generating summaries of complex research papers or trial results using summarization techniques.
Identifying Research Trends: Using topic modeling to discover emerging topics and prevalent themes within large corpora of scientific publications.
Powering Biomedical Question Answering Systems: Building systems that can answer specific questions posed by researchers or clinicians by querying large databases of scientific or clinical text.
Analyzing Patient Feedback and Social Media: Using sentiment analysis to gauge patient perception of treatments, medications, or healthcare services, potentially associated with specific entities.
Sequence Analysis: Applying techniques like autoencoders to analyze patterns or spot anomalies in biological sequences.
Conclusion
Named Entity Recognition and advanced Natural Language Processing techniques are not just technological trends; they are becoming essential capabilities for navigating the data-rich landscape of Life Sciences. By transforming unstructured text into meaningful, structured knowledge, NER and NLP accelerate research, improve patient care, and drive innovation.
While challenges related to domain specificity, ambiguity, and data sparsity exist, ongoing advancements, particularly in deep learning and Transformer models, are continually improving performance and expanding the possibilities. Leveraging these powerful tools allows researchers, clinicians, and organizations to extract hidden gems from text, gain deeper insights, and ultimately contribute to scientific discovery and better health outcomes. The journey in NLP is constantly evolving, and for Life Sciences, embracing these technologies is key to unlocking the future of biological understanding.
Athiselvam N is a skilled CapeStart Senior Software Engineer who builds scalable, high-performance apps using Spring Boot, Angular, Node.js, MySQL, MongoDB, and Elasticsearch. He specializes in microservices, APIs, LLM, OpenAI, and Agentic AI, with expertise in Python, Redis, API Gateway, AWS, and Kubernetes. Passionate about innovation, he delivers impactful solutions across domains.




